A research team led by graduate student Bryce Manifold and Assistant Professor Dan Fu has developed a new neural network architecture to interpret hyperspectral images. This new architecture incorporates deep learning based on spatial features, spectroscopy dimension, and the interplay between these two information spaces. It enables facile image segmentation, classification, and prediction of complex multidimensional hyperspectral imaging datasets.
Hyperspectral imaging encompasses a wide range of imaging techniques that not only contains two-dimensional spatial information such as photographs, but also includes an orthogonal spectral dimension. The simplest example is a color photograph, in which the color of an object reflects its absorption or scattering property. In chemistry, the spectral dimension is commonly used to distinguish the chemical composition of the object of interest. While it is extremely information-rich, disentangling and presenting object features in this multidimensional space is a challenging task. Traditional approaches rely on machine learning that segments or classifies objects based on their spectral features, while the imaging processing approach focuses heavily on spatial features.
Manifold et al. developed a new neural network, termed U-within-UNet, or UwU-Net, to address this challenge. Neural network architectures are how the “neurons” in a deep learning algorithm are connected to one another. Typical neural networks can learn the spatial features of an object, at extremely high efficiency compared to traditional methods. However, they are not built to handle hyperspectral images. UwU-Net builds off the popular U-Net to interpret spectral information and spatial information simultaneously and perform a wide variety of tasks on multiple hyperspectral imaging techniques.
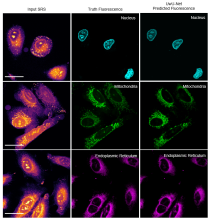
For example, the research team demonstrates the ability to classify crops and foliage from hyperspectral satellite imagery, identify anti-cancer drug locations in tissue from mass spectrometry images, and predict fluorescence of organelles in cancer cells from stimulated Raman scattering microscopy images (pictured above). One significant advantage is that the architecture is extremely flexible and can handle arbitrary spatial and spectral dimension sizes. Its wide adaptability and simple design enable many kinds of hyperspectral imaging, with spatial features from microns to miles and spectral features across all electromagnetic spectra. The team’s work is published in the March 2021 issue of Nature Machine Intelligence and can be read at https://dx.doi.org/10.1038/s42256-021-00309-y
For more information about Prof. Fu and his research, visit his faculty page or research group website.